Page 50 - Application Notebook - Solution for Food Development
P. 50
Application No.C131
News
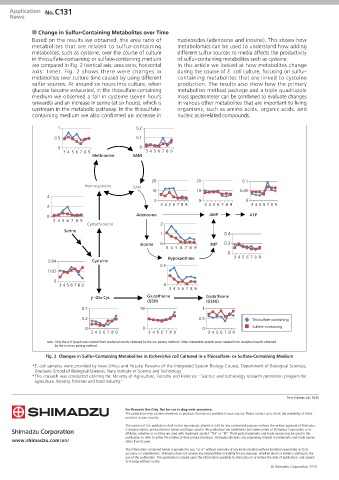
n Change in Sulfur-Containing Metabolites over Time
Based on the results we obtained, the area ratio of nucleosides (adenosine and inosine). This shows how
metabolites that are related to sulfur-containing metabolomics can be used to understand how adding
metabolites, such as cysteine, over the course of culture different sulfur sources to media affects the productivity
in thiosulfate-containing or sulfate-containing medium of sulfur-containing metabolites such as cysteine.
are compared in Fig. 2 (vertical axis: area ratio, horizontal In this article we looked at how metabolites change
axis: time). Fig. 2 shows there were changes in during the course of E. coli culture, focusing on sulfur-
metabolites over culture time caused by using different containing metabolites that are linked to cysteine
sulfur sources. At around six hours into culture, when production. The results also show how the primary
glucose became exhausted, in the thiosulfate-containing metabolites method package and a triple quadrupole
medium we observed a fall in cysteine (seven hours mass spectrometer can be combined to evaluate changes
onwards) and an increase in serine (at six hours), which is in various other metabolites that are important to living
upstream in the metabolic pathway. In the thiosulfate- organisms, such as amino acids, organic acids, and
containing medium we also confirmed an increase in nucleic acid-related compounds.
1 0.2
0.5 0.1
0 0
3456789 3456789
Methionine SAM
20 20 0.1
Homocysteine SAH
10 10 0.05
4
0 0 0
2 3456789 3456789 3456789
0 Adenosine AMP ATP
3456789
Cystathionine 2
Serine
1 0.4
Inosine 0 IMP 0.2
3456789
0
Hypoxanthine 3456789
0.04 Cysteine
0.5
0.02
0
3456789 0
3456789
γ-Glu-Cys Glutathione Glutathione
(GSH) (GSSG)
0.4 50 1
0.2 0.5 Thiosulfate-containing
0 0 0 Sulfate-containing
3456789 3456789 3456789
note: Only the ATP graph was created from analytical results obtained by the ion pairing method. Other metabolite graphs were created from analytical results obtained
by the non-ion pairing method.
Fig. 2 Changes in Sulfur-Containing Metabolites in Escherichia coli Cultured in a Thiosulfate- or Sulfate-Containing Medium
*E. coli samples were provided by Iwao Ohtsu and Yusuke Kawano of the Integrated System Biology Course, Department of Biological Sciences,
Graduate School of Biological Sciences, Nara Institute of Science and Technology.
*This research was conducted utilizing the Ministry of Agriculture, Forestry and Fisheries' "Science and technology research promotion program for
agriculture, forestry, fisheries and food industry."
First Edition: Jul. 2016
For Research Use Only. Not for use in diagnostic procedure.
This publication may contain references to products that are not available in your country. Please contact us to check the availability of these
products in your country.
The content of this publication shall not be reproduced, altered or sold for any commercial purpose without the written approval of Shimadzu.
Company names, product/service names and logos used in this publication are trademarks and trade names of Shimadzu Corporation or its
affiliates, whether or not they are used with trademark symbol “TM” or “®”. Third-party trademarks and trade names may be used in this
publication to refer to either the entities or their products/services. Shimadzu disclaims any proprietary interest in trademarks and trade names
www.shimadzu.com/an/ other than its own.
The information contained herein is provided to you "as is" without warranty of any kind including without limitation warranties as to its
accuracy or completeness. Shimadzu does not assume any responsibility or liability for any damage, whether direct or indirect, relating to the
use of this publication. This publication is based upon the information available to Shimadzu on or before the date of publication, and subject
to change without notice.
© Shimadzu Corporation, 2016