Page 8 - Oligonucleotide Therapeutics Solution Guide
P. 8
Characteristic analysis
Purification
Quality Control
Separation of Impurities Nexera XS inert
Features
Oligonucleotide Analysis by Ion Pair
Getting the right analytical conditions requires a lot of tests and data processing. Nexera XS inert can provide some great support by configuring Modification
Reverse Phase Chromatography several systems that are suitable for each purpose. Method Scouting System is a method development system based on Shimadzu’s UHPLC Target selection
technology. The combination with Method Scouting Solution dedicated control software achieves a fast and accurate method scouting workflow,
offering excellent support for method development.
Automates Development of Analytical Conditions
~ Method Scouting ~
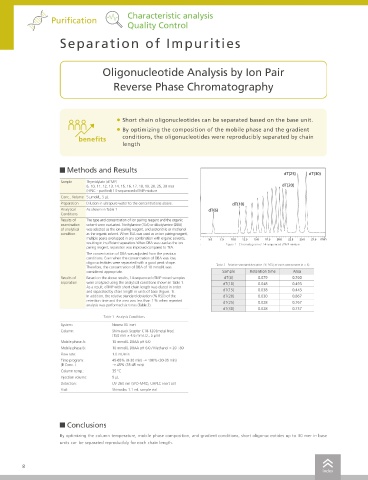
• Short chain oligonucleotides can be separated based on the base unit.
• By optimizing the composition of the mobile phase and the gradient The process of considering which analytical conditions to specify for LC separation requires evaluating a huge number of possible combinations of
columns, mobile phases, column temperatures, and other factors, which can be taxing on analytical personnel. The Nexera method scouting system
benefits conditions, the oligonucleotides were reproducibly separated by chain offers functionality for automatically switching between multiple mobile phase conditions and columns for analysis. Consequently, it can be used to
length develop methods more efficiently by automating the process of optimizing mobile phase pH, salt concentration, or other analytical condition settings. Unprotected Excision
Previous Method Oligomer synthesis
Methods and Results Create method Create schedule Acquire data Analyze data
dT(25) dT(30)
Sample Thymidylate (dTMP) A method must be created for each combination of column
6, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 25, 30 mer dT(20) and mobile phase. Creating this large number of methods is
(HPLC - purified) 14-sequenced dTMP mixture labor-intensive. Column 1 Column 2 Overnight Column 3 Column 4 Overnight Column 5 Column 6
The operation is complicated because a large amount
Conc., Volume 5 µmol/L, 5 µL of data is manually combined into one report.
Preparation Dilution in ultrapure water to the concentrations above. dT(10)
Analytical As shown in Table 1 dT(6)
Conditions Method Scouting System + LabSolutions MD
Results of The type and concentration of ion pairing reagent and the organic Create schedule Purification
examination solvent were evaluated. Triethylamine (TEA) or dibutylamine (DBA) Set conditions automatically Acquire data Analyze data
of analytical was selected as the ion-pairing reagent, and acetonitrile or methanol
condition as the organic solvent. When TEA was used as an ion pairing reagent, Methods are automatically created with
multiple peaks overlapped in any combination with organic solvents, 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 25.0 27.5 min different combinations of columns, mobile
resulting in insufficient separation. When DBA was used as the ion Figure 1 Chromatogram of 14-sequenced dTMP mixture phases, etc. Column 1 Column 2 Column 3 Column 4 Column 5 Column 6 Templates are output after Print out as a
pairing reagent, separation was improved compared to TEA.
being automatically filled 1-page report.
The concentration of DBA was adjusted from the previous with analytical results.
conditions. Even when the concentration of DBA was low,
oligonucleotides were separated with a good peak shape. Table 2 Relative standard deviation (% RSD) of each component (n = 6)
Therefore, the concentration of DBA of 10 mmol/L was mAU
considered appropriate. Sample Retention time Area 2
pH 5.7
Results of Based on the above results, 14-sequenced dTMP mixed samples dT(6) 0.079 0.760 85% 0
separation were analyzed using the analytical conditions shown in Table 1. dT(10) 0.048 0.493 A
As a result, dTMP with short chain length was eluted in order 10% B 2 pH 6.0
and separated by chain length in units of base (Figure. 1). dT(15) 0.038 0.443 X
In addition, the relative standard deviation (% RSD) of the dT(20) 0.030 0.867 5% C 0 Quality Control
retention time and the area was less than 1 % when repeated dT(25) 0.028 0.767 0% D Solvent delivery 2 Characteristic analysis
analysis was performed six times (Table 2). unit 1 pH 6.3
dT(30) 0.028 0.757 0
50% A Mixer
Table 1 Analysis Conditions 2
20% B pH 6.6
System: Nexera XS inert 5% C Y 0
Column: Shim-pack Scepter C18-120 [metal free] 25% Solvent delivery 2 pH 6.9
(150 mm × 4.6 mm I.D., 5 µm) D unit 2
0
Mobile phase A: 10 mmol/L DBAA pH 6.0 Solvent Blending Binary Gradient
Mobile phase B: 10 mmol/L DBAA pH 6.0 / Methanol = 20 : 80 2 pH 7.2
Mobile phases prepared Gradient analysis DDS
Flow rate: 1.0 mL/min 0
automatically at each pump with mobile phases
Time program: 45-65% (0-30 min) → 100% (30-35 min) 0.0 2.5 5.0 7.5 min
(B Conc. ) → 45% (35-45 min) Pharmacokinetics
Column temp.: 35 °C
Injection volume: 5 µL
Detection: UV 260 nm (SPD-M40), UHPLC inert cell Mobile Phase pH Monitor
Vial: Shimadzu 1.1 mL sample vial
~ pHM-40 ~
The pH monitor pHM-40 continuously monitors the pH of mobile phases to identify any changes in mobile
phase pH in real time.
Conclusions Other
By optimizing the column temperature, mobile phase composition, and gradient conditions, short oligonucleotides up to 30 mer in base
units can be separated reproducibly for each chain length.
8 9
index index