Page 22 - Pharmaceutical- Guide to Biopharmaceutical
P. 22
Characterization Quality Control
Protein Primary Structure Analysis PPSQ-51A / 53A + MALDI-8020
Accurate Peptide N-Terminal Amino Acid Sequencing Using benefits
a MALDI-TOF MS Mass Spectrometer and Protein Sequencer Cell Line Optimization
click here • Obtain complete sequence coverage using PPSQ and MALDI-TOF MS systems in combination.
• Enables more reliable and accurate amino acid sequencing.
Operating Principle and Features Results and Conclusion
• MALDI-TOF MS enables direct analysis of cyclic peptides or peptides with blocked N-terminals.
Using the PPSQ sequencer to analyze an amino acid sequence using Large amounts of information about peptides and proteins can be
Edman degradation, as described on the previous page (p. 20), obtained from molecular weight data measured using a MALDI-TOF
involves analyzing each amino acid one at a time, starting at the MS system. The molecular weight data is useful for quickly judging Culture
N-terminal. That eliminates mass or database dependence and other incorrect amino acid compositions and the presence of potential
problems, but Edman degradation is not well suited to processing degradations or modifications. The accurate average molecular weight
information for long sequences due to decreased reaction efficiency. of peptides can be determined easily by selecting an appropriate
To achieve more accurate and reliable N-terminal amino acid sequence matrix (Table 1). Even when using the MALDI-8020, a simple dedicated
information, combine Edman degradation data with In Source Decay linear mode system, mass is detected precisely within 20 ppm of the
(ISD) results obtained using a MALDI-TOF MS system. theoretical molecular weight.
Amino acid sequencing by mass spectrometry involves using the As shown in Table 2, N-terminal amino acid sequencing by either
differences between fragment ion masses to determine the amino acid MALDI-TOF MS or Edman degradation provides a significant benefit
sequence of peptides. ISD increases the laser output to destabilize the for identifying amino acid sequences. Of all the methods currently
substance being analyzed and break it into fragments. That results in available, N-terminal amino acid sequencing by Edman degradation Purification
obtaining a variety of fragments cleaved at the N-C α bond in peptides remains the best method for determining the actual N-terminals of
(typically C-ions). Based on the data obtained, amino acid sequences proteins and peptides. ISD also provides a reliable means of obtaining
are determined by either searching a database or by De novo sequence information, but matrix interference generally prevents
sequencing. Database searching involves comparing the measured it from being used to observe low-mass fragments relevant to
mass values to the database, which is the quickest and easiest method, N-terminals. Fig. 2 shows results from BNP analysis using a combination
but results depend on the data included in the database. In contrast, of PPSQ and MALDI-8020 systems. Only a portion of the sequence
De novo sequencing does not use a database, though it does involve can be determined using either one of these methods, but accurate
complicated data analysis that requires experience and proficiency. sequence information can be obtained for the entire length by using
Therefore, using software such as Mass++ can be helpful, because it both in a complementary way.
eliminates the need to analyze data manually. Characterization
Measurement Method Table 1 Theoretical and Measured Masses for BNP PPSQ-51A/53A Gradient System MALDI-8020
Peptide Expected mass Measured mass Mass accuracy
+
+
(ppm)
[MH+]
[MH+]
B-type natriuretic peptide (BNP), a diuretic and vasodilatory hormone BNP 5038.6 5038.5 20
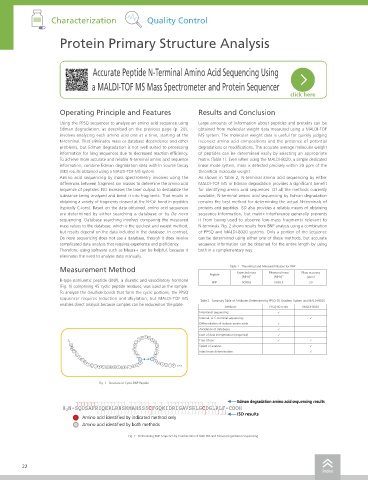
(Fig. 1) comprising 45 cyclic peptide residues, was used as the sample. Specifications
To analyze the disulfide bonds that form the cyclic portions, the PPSQ
sequencer requires reduction and alkylation, but MALDI-TOF MS Table 2 Summary Table of Attributes Determined by PPSQ-50 Gradient System and MALDI-8020 Instrument MALDI-8020
enables direct analysis because samples can be reduced on the plate.
Attribute PPSQ-50 series MALDI-8020
Mass range m/z 1 to 500,000
N-terminal sequencing
Internal, or C-terminal sequencing Mass resolution > 5,000 FWHM Quality Control
Differentiation of isobaric amino acids
Avoidance of databases Sensitivity > 250 amol
Ease of data interpretation (sequence) Mass accuracy < 20 ppm with internal calibration, < 150 ppm with external calibration
Ease of use
Acceleration voltage 15 kV
Speed of analysis
Intact mass determination Laser Solid-state laser
Wavelength 355 nm
Repetition frequency 50, 100, or 200 Hz (variable)
Flight distance 850 mm Pharmacokinetics
Fig. 1 Structure of Cyclic BNP Peptide
Detector Electron Multiplier
Ion source cleaning Includes automatic cleaning functionality (depending on built-in solid-state laser)
Edman degradation amino acid sequencing results Operating noise < 55 dB
ISD results Main unit power supply Single-phase 120 to 230 V AC, 50/60 Hz, 1,500 VA max
Amino acid identified by indicated method only Dimensions W 600 mm × D 745 mm × H 1,055 mm (excluding protrusions)
Amino acid identified by both methods Others
Weight 86 kg
Fig. 2 Determining BNP Sequence by Combination of Both ISD and Edman Degradation Sequencing Operating environment Temperature: 18 to 28 °C Humidity: Max. 70 % (with no condensation)
Note: Refer to page 21 for details of PPSQ.
22 23
index index