Page 10 - Pharmaceutical M5 Biopharma
P. 10
10 Sponsored Feature
Peptides In-depth characterization of primary structure
During process development and
before entering clinical trials, the primary
structure of a recombinant protein must be
confirmed, and all potential PTMs must be
characterized to assess the associated risk.
Peptide mapping is the fundamental
technique for this purpose, whereby the
recombinant protein is enzymatically
digested into peptide fragments that are
chromatographically separated to give
a fingerprint of the primary structure.
When coupled to mass spectrometry,
peptide mapping can also give precise
identification of various PTMs, including
oxidation, deamidation, disulfide bond
scrambling, C-terminal lysine truncation
and N-terminal pyroglutamination.
Shimadzu offers a comprehensive
portfolio of solutions for the highly
accurate confirmation of protein sequence,
identification of modifications, and routine
protein fingerprint monitoring for QA/
QC, using Part 11 compliant liquid
chromatography and LC/MS systems.
Purified Enzymatic
Protein Digestion
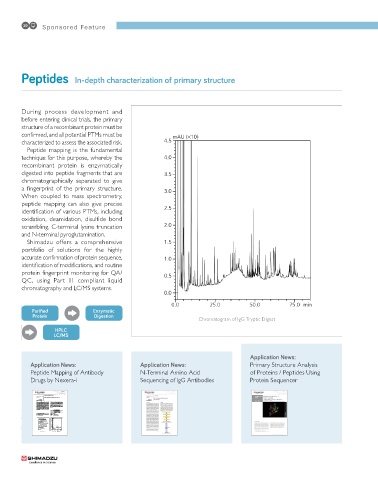
Chromatogram of IgG Tryptic Digest
HPLC
LC/MS
Application News:
Application News: Application News: Primary Structure Analysis
Peptide Mapping of Antibody N-Terminal Amino Acid of Proteins / Peptides Using
Drugs by Nexera-i Sequencing of IgG Antibodies Protein Sequencer
LAAN-A-LC-E265
Application High Performance Liquid Chromatography
News Peptide Mapping of Antibody Drugs by Nexera-i
No.L488
Peptide mapping by HPLC is one of the important quality Table 1 shows the analytical conditions. Here, the Aeris
assurance tests used for verifying the primary structure 1.7 µm PEPTIDE XB-C18 100 Å small-particle core-shell
column and the Nexera-i integrated UHPLC system was
of antibody drugs. Typically, following enzymatic
digestion of the antibodies, separation is conducted used. Mobile phase A was 0.1 % trifluoroacetic acid
using a traditional reversed phase column. Due to the (TFA) in water and mobile phase B was 0.08 % TFA in
with TFA, an optional 300 µL mixer was used.
of small-particle columns and core shell columns for acetonitrile. To ensure proper gradient performance
large number of peaks that require separation, the use
peptide analysis has spread in recent years. Fig. 2 shows the chromatogram of IgG tryptic digest, in
In order to compare elution profiles for identity and which an extremely large number of peaks are clearly
mutation confirmation, a highly repeatable system is separated.
required. The Nexera-i integrated UHPLC is the ideal
system for such an analysis. Here, the Nexera-i is used
in the analysis of IgG (human immunoglobulin G)
tryptic digest. Table 1 Analytical Conditions
Column : Aeris 1.7 µm PEPTIDE XB-C18 100 Å
(150 mm L. × 2.0 mm I.D., 1.7 µm)
Mobile Phase B: 0.08 % trifluoroacetic acid in acetonitrile
: A: 0.1 % trifluoroacetic acid in water
n Analysis of IgG Tryptic Digest Time Program Flowrate Column Temp. : B.Conc. 0 % (0 min) → 45 % (90 min)
→ 100 % (90.01 - 95 min) → 0 % (95.01 - 110 min)
: 0.2 mL/min
: 60 ˚C
For this investigation, after reduction and alkylation of
IgG, tryptic enzyme digestion was used as shown in Injection Vol. : 10 µL
Fig. 1 for sample preparation. Detection Flow Cell : High-speed high-sensitivity cell
: LC-2040C 3D at 215 nm
4.5 mAU (�10)
10 mg/mL human IgG in water 20 µL
6 mol/L guanidine hydrochloride in 4.0 3.5
0.25 mol/L Tris buffer (pH 7.5) 80 µL
0.5 mol/L dithiothreitol in water 2 µL
Incubate at 37 ˚C for 30 min 3.0
0.5 mol/L iodoacetamide in water 4.8 µL 2.5
Incubate at room temperature for 30 min in the dark 2.0
0.5 mol/L dithiothreitol in water 2 µL
0.25 mol/L Tris buffer (pH 7.5) 700 µL 1.5
1 mg/mL trypsin in 1 mmol/L HCl 4 µL 1.0
Incubate at 37 ˚C for 20 hours 0.5
Trifluoroacetic acid 1 µL
Inject to UHPLC 0.0
0.0 25.0 50.0 75.0 min
Fig. 1 Sample Preparation Fig. 2 Chromatogram of IgG Tryptic Digest