Page 19 - Microorganism Species Analysis
P. 19
MALDI-TOF MS MALDI-TOF MS
Protein Analysis (Proteome Analysis)
Microorganism Solutions Microorganism Solutions
Protein Analysis by MALDI-TOF MS Data
Peptide mass fingerprinting (PMF) is widely adopted as a proteomics approach for the easy, high-throughput analysis of
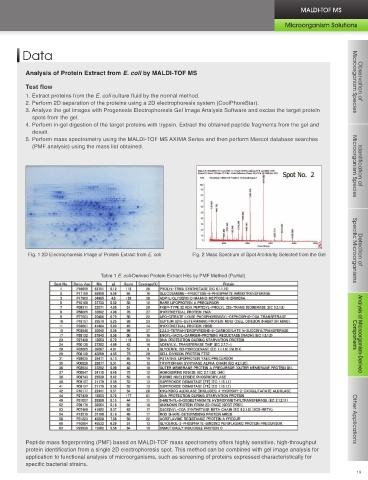
proteins using MALDI-TOF MS. With this method, after the proteins are subjected to 2D electrophoretic separation, the Analysis of Protein Extract from E. coli by MALDI-TOF MS
target proteins are enzyme digested in-gel, and a database search is performed using the mass information for the peptide Microorganism Species Observation of
fragment groups obtained to determine and identify the target proteins. Test flow
Direct amino acid sequencing (de novo sequencing) using the ORFinder-NB Mass Sequencing Kit (Protein N Terminal 1. Extract proteins from the E. coli culture fluid by the normal method.
Sequencing Kit) and protein determination by definitive amino acid sequencing with a protein sequencer (Edman reaction) 2. Perform 2D separation of the proteins using a 2D electrophoresis system (CoolPhoreStar).
Observation of
and BLAST searches are effective for handling microorganism-derived proteins not included in the protein database. 3. Analyze the gel images with Progenesis Electrophoresis Gel Image Analysis Software and excise the target protein
Microorganism Species
spots from the gel.
4. Perform in-gel digestion of the target proteins with trypsin. Extract the obtained peptide fragments from the gel and
Features of Protein Analysis (Proteome Analysis) Using the desalt.
MALDI-TOF MS AXIMA Series 5. Perform mass spectrometry using the MALDI-TOF MS AXIMA Series and then perform Mascot database searches
(PMF analysis) using the mass list obtained.
The Progenesis Electrophoresis Gel Image Analysis Software simplifies quantitative comparisons between
electrophoresis gel images and the evaluation of protein spots where a difference is apparent.
The AXIMA Series offers high-throughput and high-sensitivity protein PMF analysis and MS/MS analysis. Microorganism Species Identification of
AXIMA Confidence supports MS/MS (PSD) and AXIMA Performance supports MS/MS (CID/PSD).
Identification of
Microorganism Species
Spot matching, qualification and Mass spectrometry Database analysis
quantitation of changed spots
Fig. 1 2D Electrophoresis Image of Protein Extract from E. coli Fig. 2 Mass Spectrum of Spot Arbitrarily Selected from the Gel Specific Microorganisms Detection of
Detection of
Table 1 E. coli-Derived Protein Extract Hits by PMF Method (Partial)
Specific Microorganisms
Progenesis (2D electrophoresis) MASCOT (Matrix Science)
Progenesis SameSpots Mascot Database Search
Electrophoresis Gel Image AXIMA Performance Software (manufactured by
Analysis Software (MS/MS:CID/PSD) Matrix Science Ltd.)
Laser Desorption Ionization
Time-of-Flight Mass Spectrometer Components Analysis of Microorganism-Derived
Components
Related products
Protein N Terminal Sequencing Kit Related products
Analysis of Microorganism-Derived
ORFinder-NB Mass Sequencing Kit PPSQ-31A/33A Other Applications
AXIMA Confidence Protein Sequencing System Peptide mass fingerprinting (PMF) based on MALDI-TOF mass spectrometry offers highly sensitive, high-throughput
Other Applications
(MS/MS:PSD) protein identification from a single 2D electrophoresis spot. This method can be combined with gel image analysis for
Laser Desorption Ionization
Time-of-Flight Mass Spectrometer application to functional analysis of microorganisms, such as screening of proteins expressed characteristically for
specific bacterial strains.
18 19