Page 20 - Shimadzu Journal vol.9 Issue2
P. 20
Biopharmaceutical
Results and Discussion single enzyme digestion (shown in Fig. 3). Some of the short pep-
tide chains (around 3 to 4 amino acid) are found to be not covered;
Therapeutic proteins may undergo a series of modifications however, use of multiple enzymes for the digestion can improve
throughout their cellular production, upstream/downstream pro- the sequence coverage.
cessing, and storage. These modifications can include the addition LCMS-9030 offered excellent mass accuracies for the peptides
or replacement of functional groups, or structural changes such as with different chain length. Representative data demonstrating the
folding/unfolding, cleavage, and racemization. Presence of these mass accuracy (less than 2 ppm) obtained for peptides with chain
modifications can affect biological activity, half-life and immuno- length as short as 4 amino acid and as long as 63 amino acid is shown
5
genicity . Hence, it is of utmost importance to identify these mod- in Table 2. Obtaining such mass accuracies and stability for longer
ifications accurately. Moreover, it is also important to find out the duration is of utmost important to accurately identify the PTMs.
sites which are prone to undergo such modifications. Generally, mass shift in precursor ion m/z for the modified pep-
Analysis of the control and artificially stress induced mAb sam- tide and change in the retention time are considered to identify the
ples can provide understanding of such susceptible sites. The bot- presence of PTMs. Furthermore, acquiring good quality MS/MS
tom-up approach for mAb characterization is typically referred to spectra is equally important to confidently assign the location of the
as “peptide mapping,”. Peptide mapping analysis not only provides modification on a given peptide. Collision energy spread function
information about primary sequence of mAbs but also useful in of LCMS-9030 helps acquire MS/MS fragmentation pattern over a
identifying sites that are susceptible to oxidation, deamidation etc. range of collision energy (18-52 V, in this case) instead of obtaining
Hence, control and stress induced samples of trastuzumab bi- MS/MS spectra at single or few selected collision energies. Thus,
osimilar were subjected to peptide mapping and PTMs analysis. comprehensive MS/MS fragmentation pattern can be obtained
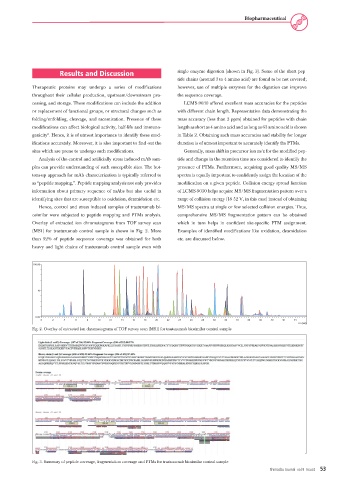
Overlay of extracted ion chromatograms from TOF survey scan which in turn helps in confident site-specific PTM assignment.
(MS1) for trastuzumab control sample is shown in Fig. 2. More Examples of identified modifications like oxidation, deamidation
than 92% of peptide sequence coverage was obtained for both etc. are discussed below.
heavy and light chains of trastuzumab control sample even with
100.00
%
0.00
0 2 4 6 8 10 12 14 16 18 20 22 24 26 28 30 32 34 36 38 40 42 44
RT (min)
Fig. 2. Overlay of extracted ion chromatograms of TOF survey scan (MS1) for trastuzumab biosimilar control sample
Fig. 3. Summary of peptide coverage, fragmentation coverage and PTMs for trastuzumab biosimilar control sample
53
Shimadzu Journal vol.9 Issue2 53