Page 15 - LifeScience Solution for Lipid and Lipidome
P. 15
Structural Analysis of Glycolipids
Data Subcutaneous Tissue Mass Imaging of
Glycolipids are molecules with a sugar chain bonded to a lipid, where
the lipid portion interacts with biological membranes and the sugar HO OH HO OH
OH O O
Ganglioside is the general term for sphingoglycolipids containing sialic acid, which are concentrated in lipid rafts on the cell portion protrudes outside the cell. Sugar chains are recognized by sugar HO OH O O O
membrane surface. They are presumed to be involved in modulating cell signal transduction, but many aspects of their role are still chain-recognizing molecules and serve a variety of biological functions, HO O OH O NH O OH O OH Analysis of
not understood. such as cell-cell recognition, signal transduction, bacterial and viral HO OH O OH O O Comprehensive
O N HO O HO O
Fig. 1 shows the structures of “GM1”, “GD1b”, and “GalNAc-GM1b”. adhesion, and bonding with toxins. Until now, the structure of sugar H O OH OH Glycerophospholipids
O HO
Fig. 2 shows the results from investigating the composition of gangliosides in T-cell subsets that serve an important role in immune chains could be predicted in minute quantities using antibodies or H OH
response. lectin, a sugar chain-recognizing molecule. However, with almost no N H HO
Differences in the content of gangliosides between each cell, expressed as a proportion of the added internal standard substance way to predict the structure of the lipid portion, glycolipids with NH H
+
(ISD), can be investigated quantitatively. A dramatic increase in GalNAc-GM1b and extended GM1b in CD8 T-cells is evident. At uniform sugar chains and a mixture of different lipids were commonly
the same time, there is a remarkable increase in molecules with ceramide structures d18:1-24:1, d18:1-22:0, and d18:1-24:0. used. Using an LCMS-IT-TOF system allows identification of not only the O
+
+
There is a higher proportion of GD1 in CD4 T-cells than in CD8 T-cells. This pattern is close to that of thymocytes, but the sugar chain structure, but also the structure of the lipid portion, even in Technology
proportion of d18:1-22:0 and d18:1-24:0 is higher. Future research issues include differences in the expression level of such mixtures containing trace quantities of biological samples. GD1a(d18:1-16:0) Supercritical Fluid Lipid Analysis Using
gangliosides and cell differentiation as well as the relationship with the associated differentiation of their functions.
OH
HO OH
HO
OH HO OH O O
O O NH
HO OH HO
HO O O O OH O HO O OH HO
OH NH OH O OH HO O O OH O
O O O O O
HO OH O OH O HO OH NH OH O OH HO O O OH O Analysis of
O HO O O O O NH OH
OH OH HO HO O O O OH O O HO O HO HO O O OH O
O HO O HO OH OH O OH HO OH O HO OH O
H OH HO HO O NH Glycerophospholipids
HO H
N HO NH O OH
H O N H OH
H HO
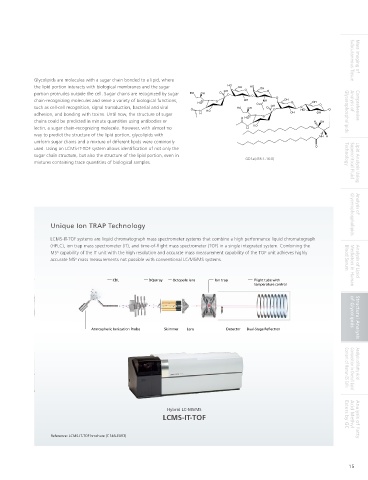
NH H NH H NH H Unique Ion TRAP Technology
O O O
GM1 (d18:1-16:0) GD1b (d18:0-18:0) GalNAc-GM1b (d18:0-16:0) LCMS-IT-TOF systems are liquid chromatograph mass spectrometer systems that combine a high performance liquid chromatograph
(HPLC), ion trap mass spectrometer (IT), and time-of-flight mass spectrometer (TOF) in a single integrated system. Combining the
Fig. 1 Structural Formula of GM1, GD1b, and GalNAc-GM1b
MS capability of the IT unit with the high resolution and accurate mass measurement capability of the TOF unit achieves highly
n
n
accurate MS mass measurements not possible with conventional LC/MS/MS systems. Blood Serum Analysis of Lipid
+
+
Thymocytes CD4 T cells CD8 T cells Mediators in Human
(×1,000,000) (×1,000,000)
2.75 1.1 (×1,000,000)
2.50 1.0 1.2
2.25 24:1 0.9 ISD 24:1 1.1 1.0 ISD 24:1 22:0
2.00 ISD 16:0 0.8 0.9
1.75 0.7 16:0 22:0 0.8 CDL DQarray Octopole lens Ion trap Flight tube with
1.50 0.6 0.7 16:0 24:0
GM1 1.25 0.5 0.4 18:0 20:0 24:0 0.6 0.5 20:0 temperature control
1.00
0.75 22:0 24:0 0.3 0.4 0.3 18:0
0.50 0.2 0.2
0.25 0.1 0.1
0.00 0.0 0.0
0.0 2.5 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 25.0 27.5 30.0 32.5 35.0 37.5 0.0 2.5 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 25.0 27.5 30.0 32.5 35.0 37.5 0.0 2.5 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 25.0 27.5 30.0 32.5 35.0 37.5
(×1,000,000) (×1,000,000) (×1,000,000)
2.25 1.2
2.00 24:1 1.1 1.0 ISD 24:1 1.1 ISD
1.75 ISD 16:0 0.9 16:0 1.0
1.50 0.8 0.7 22:0 0.9 0.8 of Glycolipids
24:1
1.25 22:0 24:0 0.6 0.5 0.4 0.3 0.2 18:0 20:0 24:0 0.7 0.6 0.5 0.4 0.3 16:0 18:0 20:0 22:0 24:0 Structural Analysis
Relative intensity 0.25 0.0 (×1,000,000) ISD 7.5 10.0 12.5 15.0 17.5 20.0 22.5 25.0 27.5 24:1 32.5 35.0 37.5 1.75 0.1 0.0 0.0 (×1,000,000) 5.0 7.5 10.0 16:0 15.0 17.5 20.0 22.5 25.0 27.5 30.0 22:0 35.0 37.5 0.2 0.1 0.0 4.5 4.0 3.5 0.0 (×1,000,000) 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 24:1 22:0 35.0 37.5 Atmospheric Ionization Probe Skimmer Lens Detector Dual-Stage Re ectron
GD1
1.00
0.75
0.50
0.00
32.5
12.5
2.5
30.0
25.0
27.5
2.5
32.5
30.0
2.5
5.0
2.00
24:1
1.75
1.50
24:0
1.50
GalNAc-
1.25
1.25
1.00
GM1b
0.75
0.75
0.50
0.50 16:0 22:0 24:0 1.00 ISD 18:0 20:0 24:0 3.0 2.5 2.0 1.5 1.0 ISD 16:0 18:0 20:0
0.25 0.25 0.5
0.00 0.00 0.0
0.0 2.5 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 25.0 27.5 30.0 32.5 35.0 37.5 0.0 2.5 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 25.0 27.5 30.0 32.5 35.0 37.5 0.0 2.5 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 25.0 27.5 30.0 32.5 35.0 37.5
(×1,000,000) (×1,000,000) Analysis of Fatty Acid
(×1,000,000)
2.00 1.3 4.5
1.2 24:1 Content of Human ES Cells
1.75 1.1 24:1 4.0 22:0 Composition in Overall Lipid
1.50 ISD 1.0 0.9 ISD 3.5 3.0
extended 1.25 0.8 0.7 0.6 22:0 2.5 20:0 24:0
1.00
GM1b 0.75 24:1 0.5 16:0 20:0 24:0 2.0 1.5 ISD 16:0
0.50 0.4 0.3 18:0 1.0 18:0
0.25 16:0 22:0 24:0 0.2 0.1 0.5
0.00 0.0 0.0
0.0 2.5 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 25.0 27.5 30.0 32.5 35.0 37.5 0.0 2.5 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 25.0 27.5 30.0 32.5 35.0 37.5 0.0 2.5 5.0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 25.0 27.5 30.0 32.5 35.0 37.5
Retention time Retention time Retention time
+
+
Fig. 2 Mass Chromatograms Obtained from Analysis of Gangliosides of Thymocytes, CD4 T-Cells and CD8 T-Cells Using the C30 Column
Hybrid LC-MS/MS
LCMS-IT-TOF Esters by GC Acid Methyl
This data was reported in the paper titled “CD4 + and CD8 + T-cells require different membrane gangliosides for activation”, Nagafuku M, Okuyama K, Onimaru Y, Analysis of Fatty
Suzuki A, Odagiri Y, Yamashita T, Iwasaki K, Fujiwara M, Takayanagi M, Inokuchi J (2012) Proc. Natl. Acad. Sci. U.S.A. 109, E336–E342.
Reference: Technical Report “Structural Analysis of Glycosphingolipids by LC-IT-TOF-MS” (C146-E268) Reference: LCMS-IT-TOF brochure (C146-E093)
14 15