Page 18 - Oligonucleotide Therapeutics Solution Guide
P. 18
Characteristic analysis
Quality Control
Molecular Weight Confirmation MALDI-8030
Features
Negative Mode Analysis of Synthetic Oligonucleotides using the
The MALDI-8030 is the latest in a long line of MALDI-TOF products from Shimadzu. Instrument performance specifications are extended from Modification
MALDI-8030 Dual Polarity Benchtop MALDI-TOF Mass Spectrometer those of the MALDI-8020 to cater for compounds best suited to analysis in negative ion mode. This dual-polarity, benchtop linear MALDI-TOF mass Target selection
spectrometer delivers outstanding performance in a compact footprint, making it an ideal choice for today’s increasingly demanding laboratories.
click here This high throughput and flexible analytical capabilities are ideal for quality control and profiling applications such as peptides, proteins, polymers
and oligonucleotides.
• Simple analysis of oligonucleotides, in negative-ion mode to reduce adducts,
on an affordable benchtop MALDI-TOF MS
• Dual mode (positive / negative ion) MALDI-TOF
• Quality spectra with good mass accuracy offers MALDI-TOF MS as an
benefits alternative to gel-Ethidium bromide detection post PCR • 200 Hz solid-state laser, 355 nm Excision
• Load-lock chamber for fast sample introduction Unprotected
• UV laser-based source cleaning (Pat. US 10340131) Oligomer synthesis
Methods and Results • Small footprint/benchtop design
• Quiet operation (<55 dB)
Sample 5‘-ATCTTTGGTTT-3‘ (Wild type / normal CFTR gene) Average
mass : m/z 3656.43
5'-ATTGGTGT-3‘ (Phe508delCFTR mutated gene)
Average mass : m/z 2758.85
Conc., Volume 100 µmol/L, 1 ~ 10 µL
Preparation Samples were diluted with ultrapure water to the above Sample Target Solutions
concentration.
Matrix solution Ammonium citrate dibasic was prepared at 5 mg/mL in 70:30 Compatible with the FlexiMass series of microscope slide-format sample targets, Purification
acetonitrile/water, which was used to prepare the 3-HPA matrix these slides provide researchers with options depending on their experimental
(45 mg/mL). Samples were pre-mixed with matrix (1:2) prior to workflow.
spotting onto the MALDI target.
The individually barcoded, single-use FlexiMass-DS slides provide a convenient
Positive ion Desalting was performed using the Dowex cation exchange resin, solution for more routine or defined workflows. Ready-to-use, these disposable
mode which works through exchange of hydrogens for sodium and targets eliminate the need for cleaning and the risk of carryover. Alternatively, the
other salts.
reusable stainless steel FlexiMass-SR sample targets provide a cost-effective, longer-
Negative ion No desalting was performed for the negative-ion mode analysis. term solution to sample preparation.
mode
analysis MALDI analyses were conducted on the MALDI-8030 in positive
and negative ion modes using the desalted and non-desalted
samples, respectively.
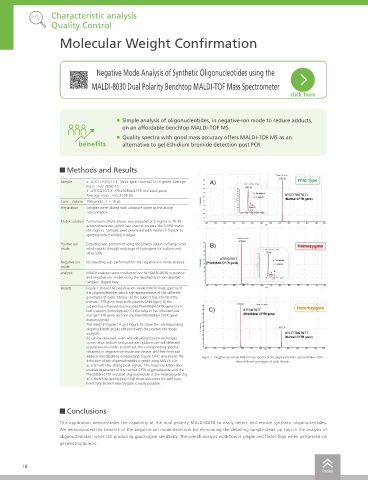
Results Figure 1 shows the negative-ion mode MALDI mass spectra of Automated Source
the oligonucleotides which are representative of the different
genotypes of cystic fibrosis: A) the subject has inherited the Cleaning TrueClean Quality Control
normal CFTR gene from both parents (wild type); B) the Characteristic analysis
subject has inherited the mutated Phe508del CFTR gene from To maintain instrument performance over time, the MALDI-8030 features wide-bore ion optics
both parents (homozygote); C) the subject has inherited one
normal CFTR gene and one mutated Phe508del CFTR gene which minimize the risk of source contamination and provide a robust platform. TrueClean, an
(heterozygote). automated source cleaning function, cleans the extraction electrode in-situ quickly within 10 min by
The insets in Figure 1A and Figure 1B show the corresponding using UV laser without breaking instrument vacuum.
oligonucleotide peaks obtained with the positive-ion mode
analyses.
As can be observed, even with desalting (cation exchange),
some minor sodium and potassium adducts are still detected
in positive-ion mode. In contrast, the corresponding spectra
obtained in negative-ion mode are cleaner and free from salt DDS
adducts (no desalting is necessary). Figure 1A-C also shows the Figure 1 Negative-ion mode MALDI mass spectra of the oligonucleotides representative of the
detection of the oligonucleotides is good using MALDI-TOF, three different genotypes of cystic fibrosis
as seen with the strong peak signals. The mass resolution also
enables separation of the normal CFTR oligonucleotide and the Pharmacokinetics
Phe508del CFTR mutated oligonucleotide in the heterozygote (Fig
4C), therefore genotyping of all three outcomes for wild type,
High Quality with Low-running Cost Database management
homozygote and heterozygote is easily possible.
For improving product performance and the longevity, components Operating under the control of MALDI Solutions software, the
were carefully selected, which realized a robust platform capable of software features a centralized, secure Microsoft SQL database which
Conclusions delivering outstanding performance in a small footprint. Simple design can be used to store everything from sample lists and acquisition
provides easy maintenance and contribute to maintaining the quality Parameter Sets to acquired MALDI data. The system is managed by
This application demonstrates the capability of the dual polarity MALDI-8030 to easily detect and resolve synthetic oligonucleotides. of analysis over time. an administrator and customizable user profiles provide control over Other
access to the database and operation of the instrument. Along with a
We demonstrated the benefits of the negative ion mode detection for eliminating the desalting sample clean up step in the analysis of
full audit trail, the software provides the tools to help achieve 21 CFR
oligonucleotides, while still producing good signal sensitivity. The overall analysis workflow is simple and faster than when performed via part 11 regulatory compliance.
gel electrophoresis.
18 19
index index